System Architecture¶
GeneInsight employs a two-stage approach to extract and organize biological information from gene sets.
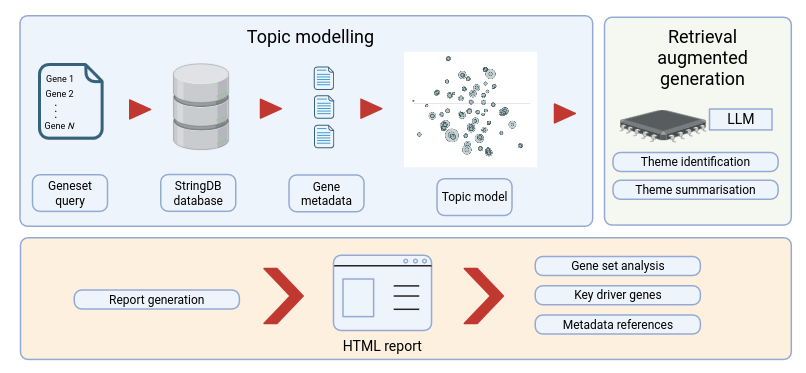
Figure 1: GeneInsight system architecture and workflow¶
Biological Theme Generation Stage¶
Annotation Collection: The system collects functional annotations from the STRING database for each input gene, creating a collection of gene-specific descriptions.
Cluster-based Topic Modeling: This textual corpus is subjected to cluster-based topic modeling, which groups similar annotations into clusters (topics) and identifies key terms for each cluster.
LLM Theme Generation: A large language model (LLM) then converts representative annotations from each cluster into interpretable biological themes.
Gene-Theme Linking: These biological themes are linked back to genes via their associated descriptions.
Statistical Validation: The system performs hypergeometric testing with false discovery rate correction to identify which biological concepts are significantly enriched within the original gene set.
Summarization Stage¶
Theme Refinement: Another round of cluster-based topic modeling identifies key themes, measuring how consistently they appear as cluster representatives across multiple runs.
Hierarchical Structuring: The software extracts the final summary by selecting themes based on user-defined length preferences.
Interactive Report Generation: A large language model creates a hierarchical summary where major biological themes appear as main headings with related subheadings grouped beneath them.
Cross-reference Integration: The final interactive HTML report seamlessly links theme descriptions to their corresponding gene annotations, enabling researchers to navigate between overarching biological processes and their specific components.
Technical Implementation¶
GeneInsight is implemented in Python (3.9+) and distributed as a Docker container to ensure reproducible deployment across platforms. The core computational components include:
BERTopic (v0.15.0) for topic modeling
SentenceTransformer for generating dense vector representations
Optuna (v3.3.0) for hyperparameter optimization
Snakemake (v7.32.4) for workflow management
The system employs multiple rounds of topic modeling with different random seeds to ensure robust theme identification, identifying stable topics through cosine similarity measurements between iterations.