GeneInsight Documentation¶



GeneInsight: AI-powered tool for gene set interpretation through advanced topic modeling and large language models.
Overview¶
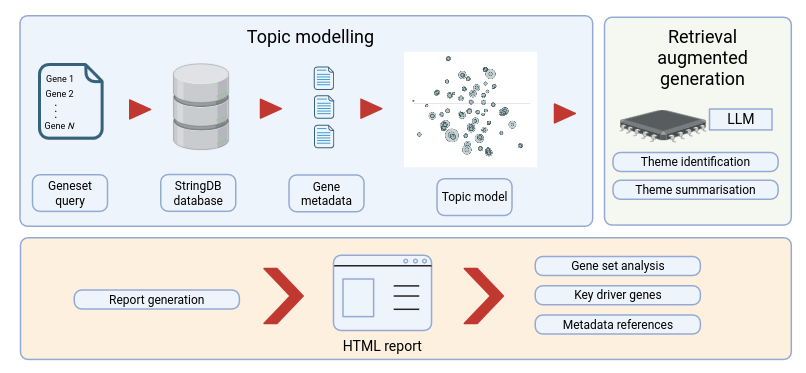
GeneInsight addresses the challenge of interpreting gene sets by combining advanced topic modeling with large language models to automatically synthesize diverse biological annotations. It consolidates extensive annotations into coherent thematic summaries, enabling rapid extraction of biologically significant insights that conventional enrichment analyses often overlook.
The tool provides a comprehensive pipeline for analyzing gene sets:
Gene Enrichment: Query STRING database (https://string-db.org/) for gene-specific annotations
Topic Modeling: Apply BERTopic (https://maartengr.github.io/BERTopic/index.html) to identify coherent biological themes
LLM Integration: Use language models to refine and interpret topics through Retrieval Augmented Generation (RAG), enhancing topic interpretations with domain-specific knowledge
Enrichment Analysis: Perform hypergeometric testing to validate biological relevance
Results Visualization: Generate interactive reports with biological insights
Key Features¶
Integrates gene-specific annotations from multiple sources including STRING database
Employs cluster-based topic modeling to identify coherent biological themes
Utilizes large language models for automated knowledge synthesis
Performs statistical validation through hypergeometric testing
Generates hierarchical summaries with interactive visualization
Supports multiple species through NCBI taxonomy IDs
Offers both command-line interface and API access
References¶
- BERTopic
https://maartengr.github.io/BERTopic/index.html - Advanced topic modeling technique that leverages BERT embeddings and clustering algorithms to create coherent topics
- STRING-DB
https://string-db.org/ - Database of known and predicted protein-protein interactions, providing comprehensive gene annotations